Introduction
- Instance Segmentation: object detection + semantic segmentation
- challenging because
- we need the correct detection of all objects in an image
- AND precisely segment each image
- Mask R-CNN: a ‘conceptually simple’, flexible, and general framework for object instance segmentation
- extension of Faster R-CNN: a parallel mask branch, Feature Pyramid Network, RoIAlign
Mask R-CNN structure overview
-
Two(+1?) main stages
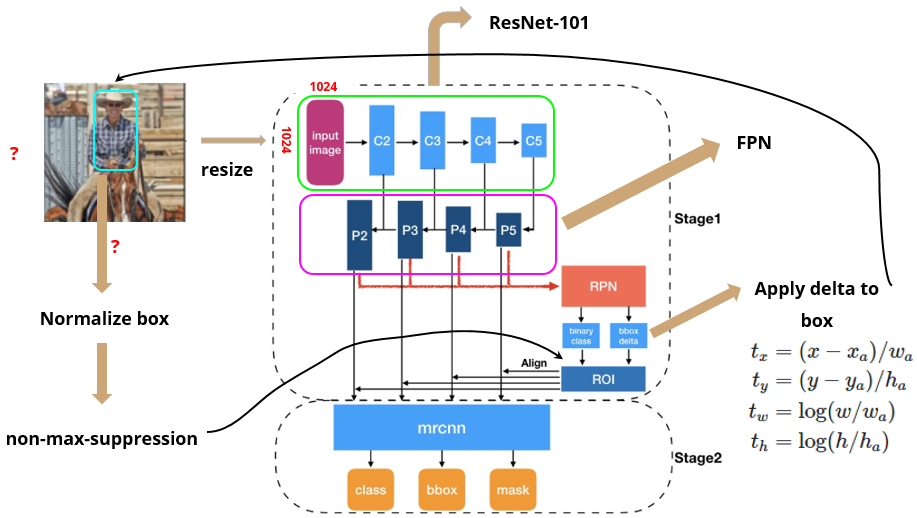
- ResNet-FPN backbone: scan image and extract features over entire image
- pass image into backbone network (ex. ResNet, VGG)
- extract feature map (via Feature Pyramid Network)
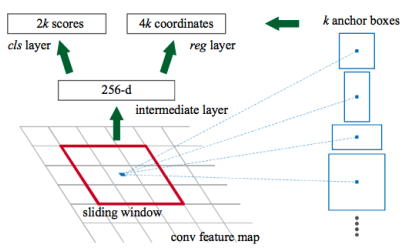
1.5. Region Proposal Network
- pass feature map to RPN (Region Proposal Network) to generate proposals (areas likely to contain an object)
- using proposals from RPN, get RoI (use Non-Max Suppression if needed)
- return fixed size feature map (via RoIAlign)
- Network head: parallel predictions of class, box offset, and binary mask for each RoI
- pass fixed size feature map from RoIAlign operations into Faster R-CNN
- ResNet-FPN backbone: scan image and extract features over entire image
Mask R-CNN: extension of Faster R-CNN
1. Feature Pyramid Network
addition of FPN (Feature Pyramid Network) before RPN (Region Proposal Network)
- Faster R-CNN: get RoI’s from 1 feature map from backbone
- only left with most important features in the end → loses intermediary features
- make anchors of various scales and orientations to detect objects of various shapes → inefficient
- FPN: get feature maps by adding previous feature maps → preserve information from previous/smaller feature maps
- no need to make anchors of various sizes with each feature map (small feature map → large anchor, vice versa)
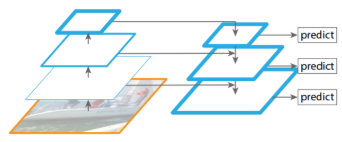
2. RoIAlign
used RoIAlign, instead of RoI pooling, for masking of image segmentation
- Mask: encodes an input object’s spatial layout
- naturally addressed by pixel-to-pixel correspondence provided by convolution (not fully connected layers, which collapse info into short output vectors)
- predicts m x m mask from each RoI using a FCN (1x1 conv) to maintain spatial dimensions
- require fewer parameters and is more accurate
- necessary assumption for pixel-to-pixel correspondence: RoI features (small feature maps) are well aligned to faithfully preserve the explicit per-pixel spatial correspondence → motivation to use RoIAlign
- Before making feature maps into a fixed size with RoIPool or RoIAlign
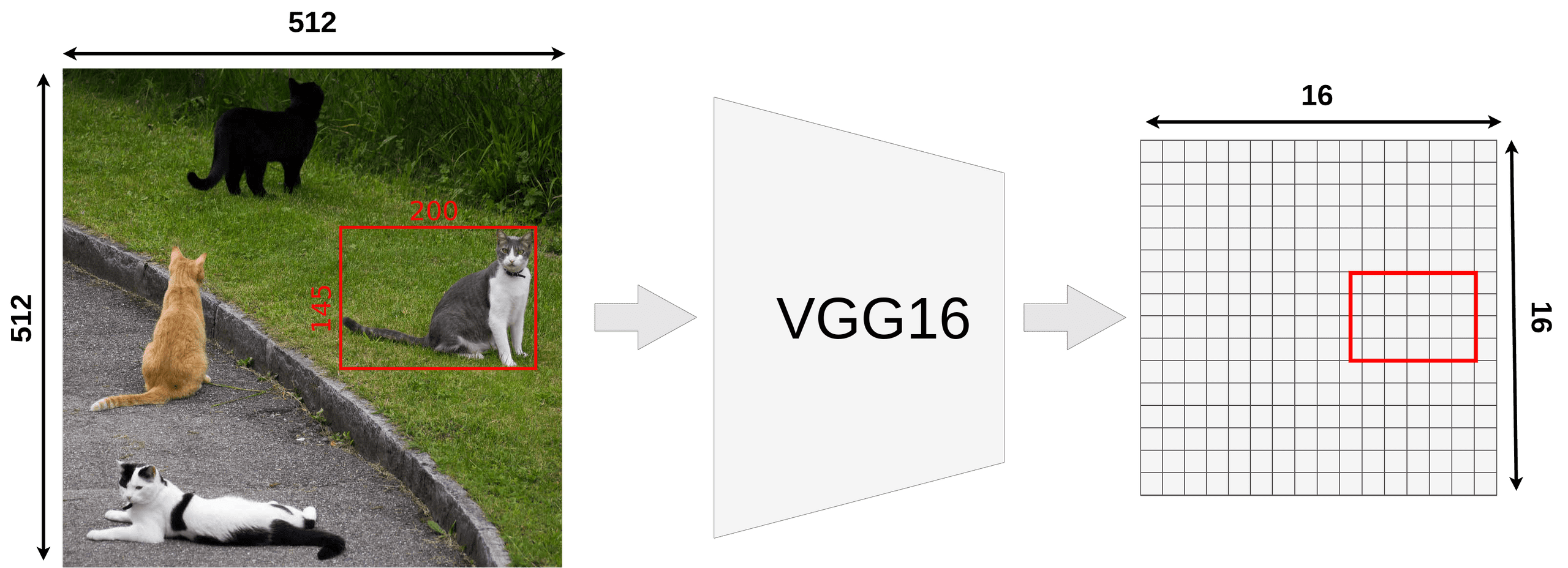
region proposals from RPN: offsets for each anchors- Each feature map (in FPN) have been decreased k times from the original image
- we want to map proposed RoI onto feature map
- not all object dimensions can be divided by k → RoI may not align with our grid for feature map
- RoI pooling performs coarse spatial quantization for feature extraction (lacks pixel-to-pixel alignment)
- Quantization of floating number RoI to discrete granularity of feature map (map FL → feature map cells?)
- Quantization: mapping input from large set → output in a smaller set. (e.g. rounding, truncation)
- Granularity: composition of distinguishable pieces
- Quantized RoI subdivided into spatial bins
- spatial bins quantized (ex. [x/16] → round after applying stride = 16)
-
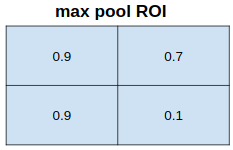
feature values covered by each bin → aggregated (ex. max pooling)
→ Quantization introduce misalignments between RoI and extracted features
→ Robust to small translations, but bad for predicting pixel-accurate masks
- Quantization of floating number RoI to discrete granularity of feature map (map FL → feature map cells?)
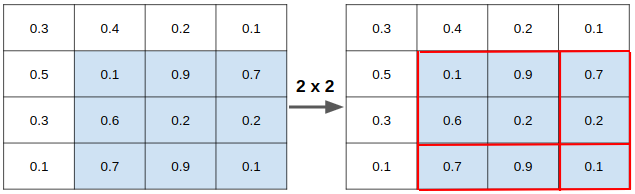
- RoIAlign: faithfully preserves exact spatial locations → simple, quantization-free layer
- RoIAlign layer removes harsh quantization of RoIPool and properly aligns extracted features with the input
- avoid any quantization of RoI boundaries or bins (→ no rounding. just divide each coordinate x by 16 instead of [x/16])
→ potentially no definite pixel grid (because new coordinates are in floats)
→ misalignment!
- Divide RoI into equal size boxes and apply bilinear interpolation inside every box (ex. 3x3 pooling layer → 9 boxes)
- get four sampling points inside each box (1/3, 2/3 지점 in x, y axis)
- results are not sensitive to exact sampling locations / # of samples (as long as there's no quantization)
- bilinear interpolation to compute exact values of input features at four regularly sampled locations in each RoI bin → aggregate results (ex. max pooling)
- bilinear interpolation → get weighted average of centers of the 4 nearest boxes
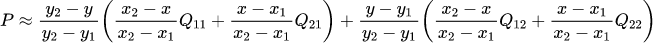
→ $Q_{ij}$: values of each box, $(x,y)$: coordinates of sampled location, $(x_i,y_i)$: coordinates of each box's center

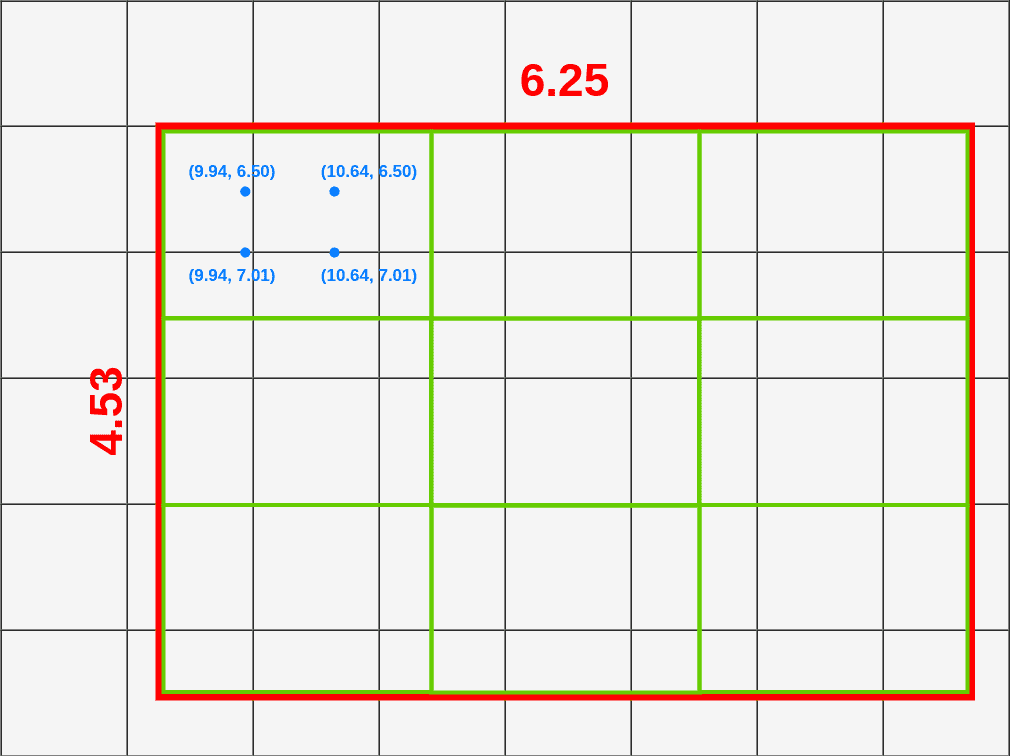
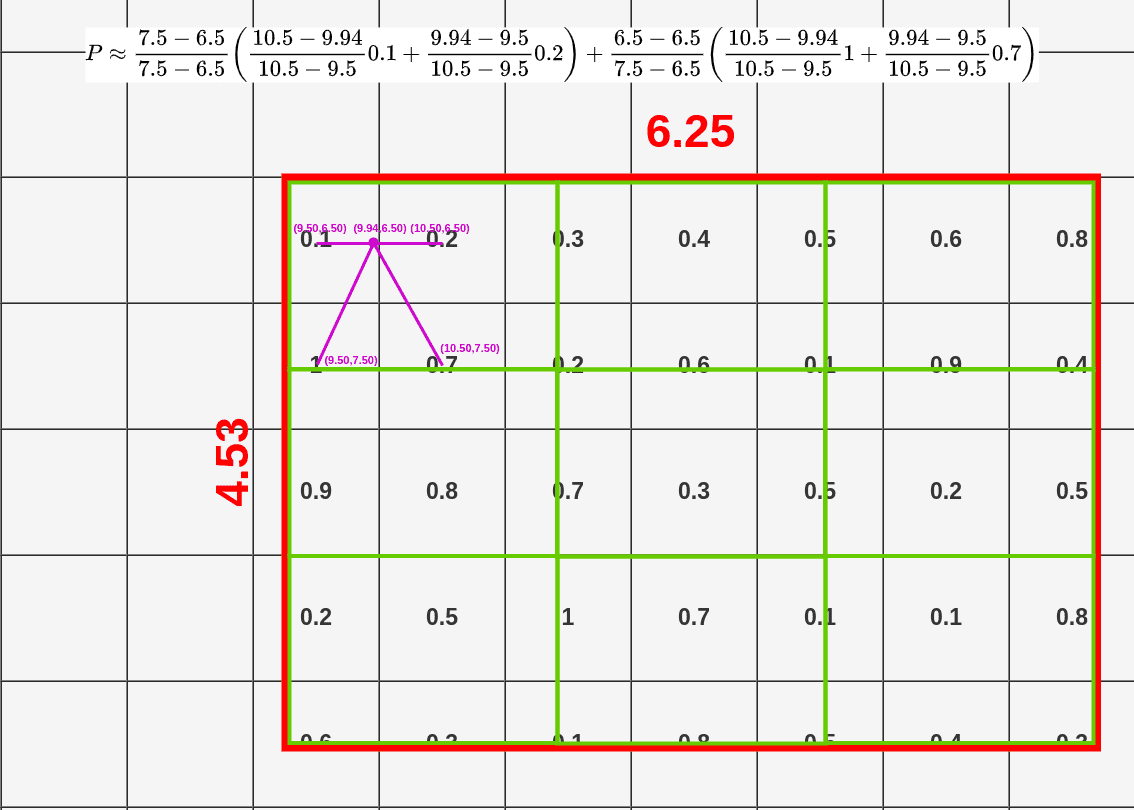
→ get weighted average for each sampled location (here, P=0.14)
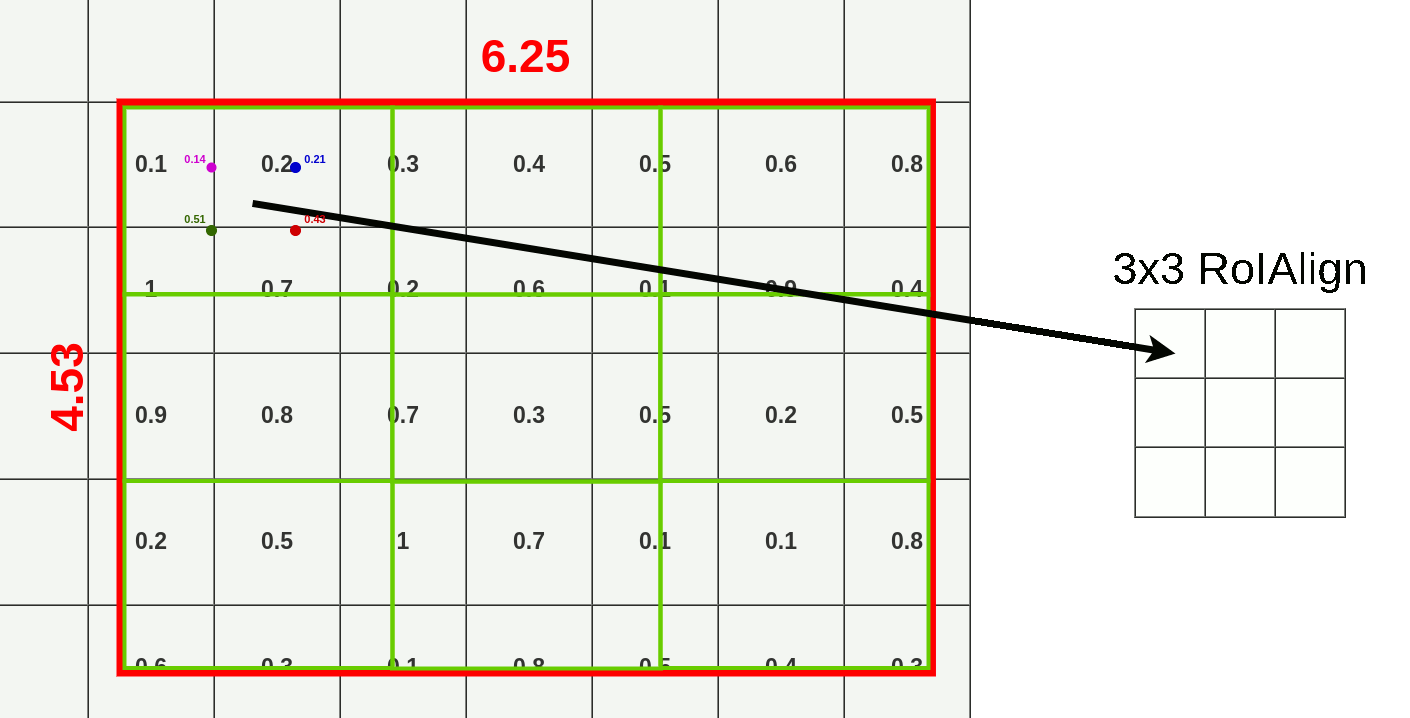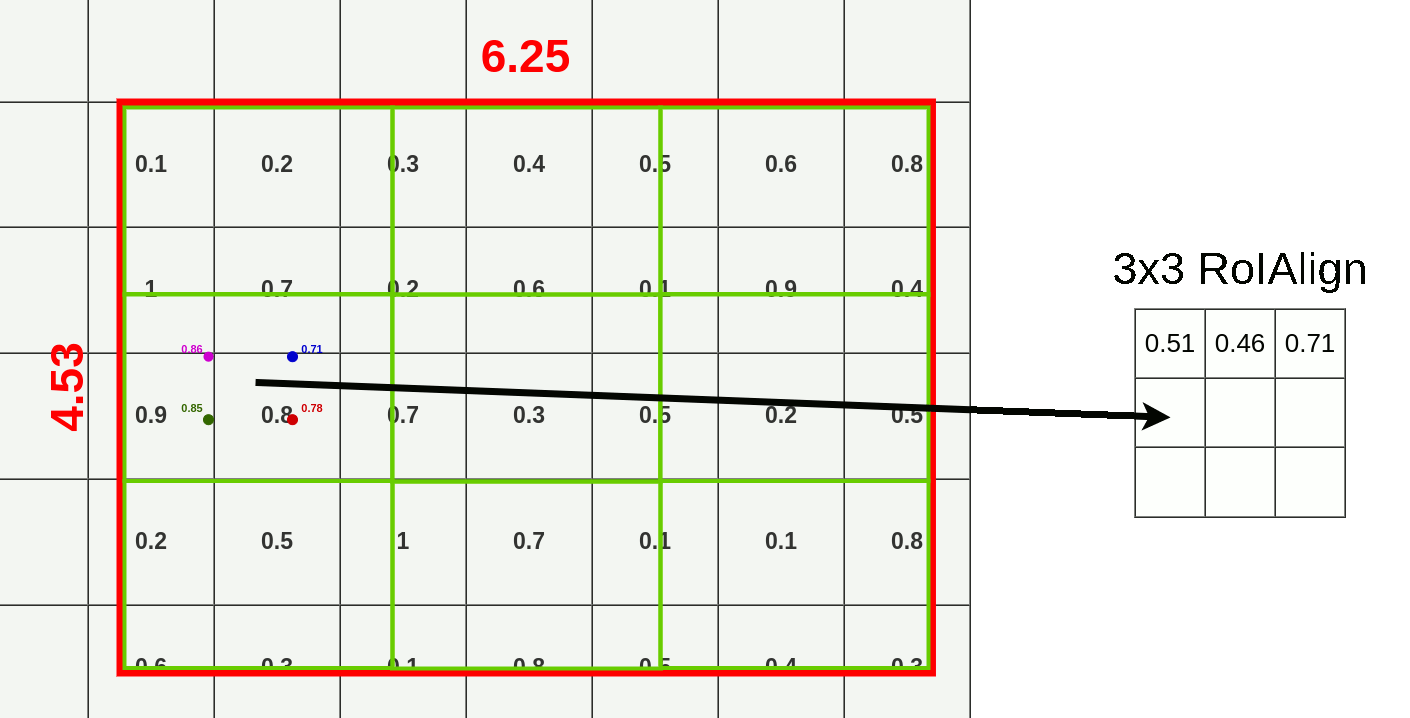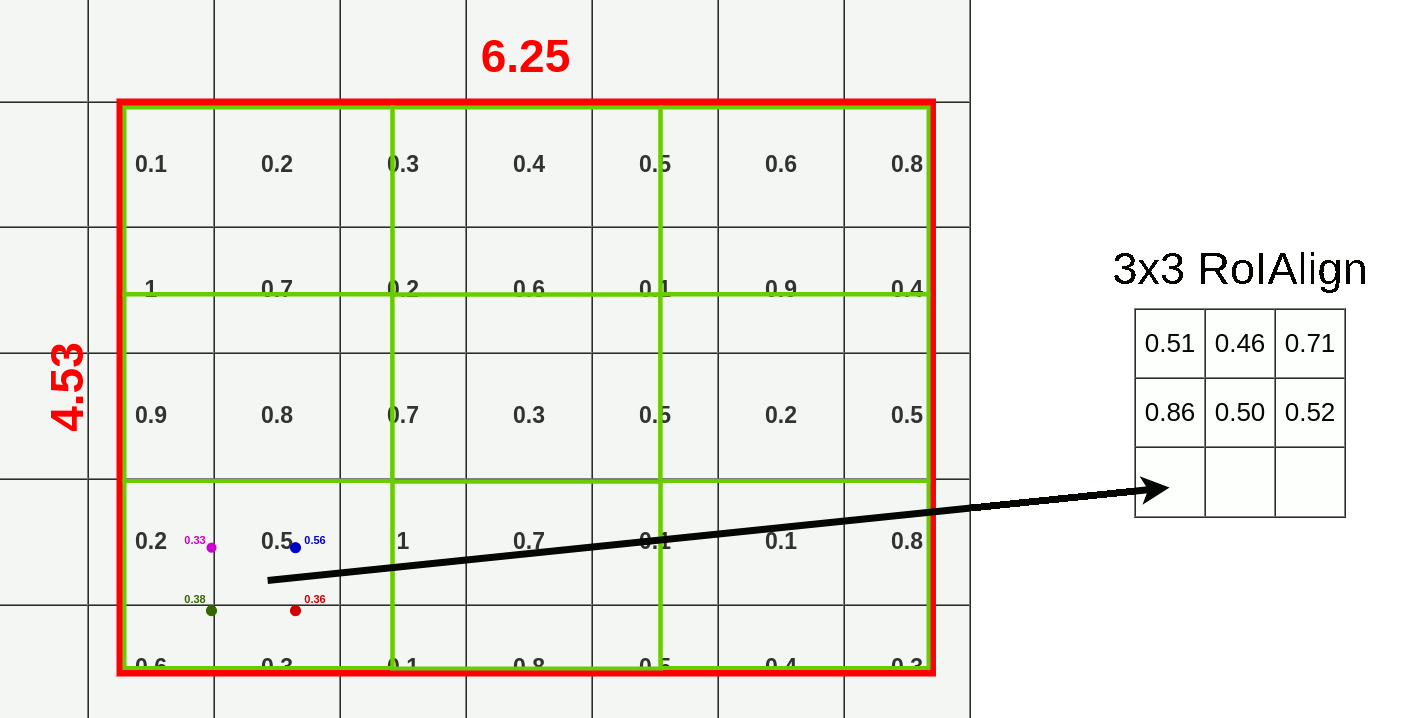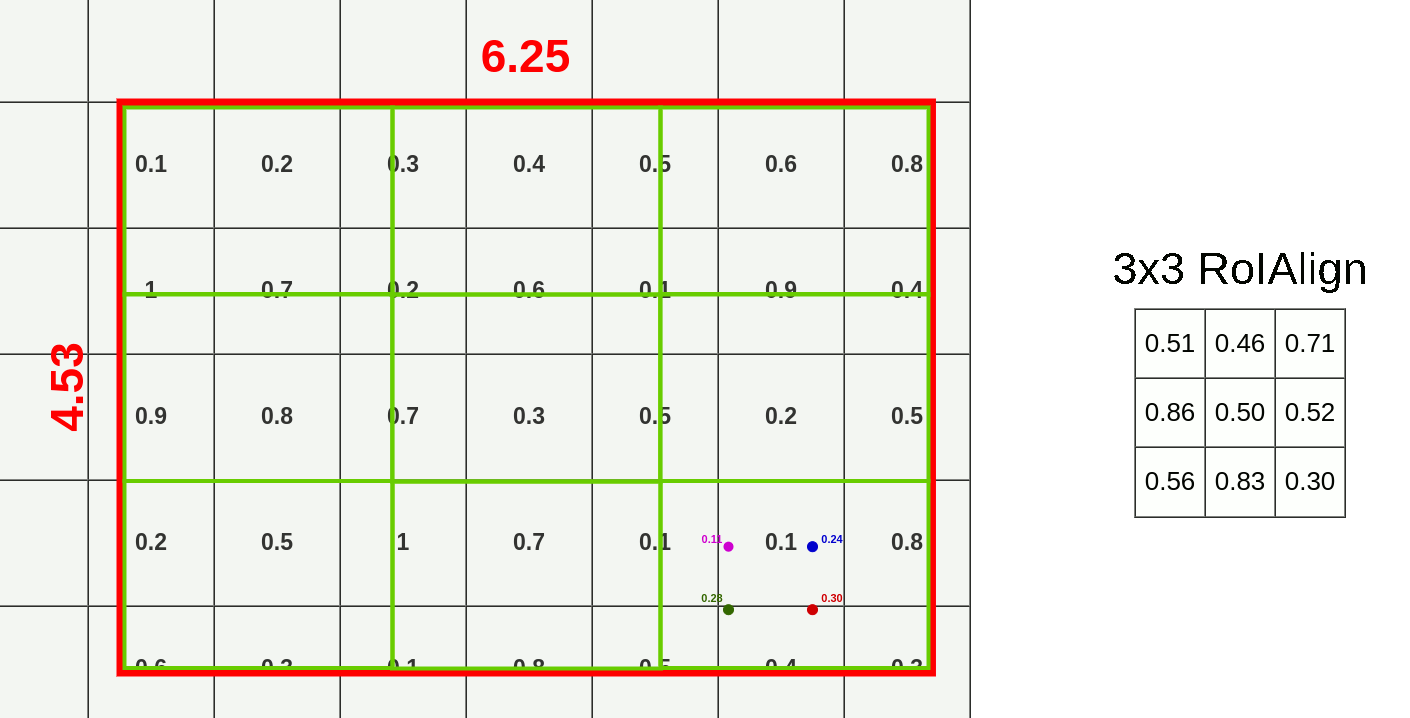
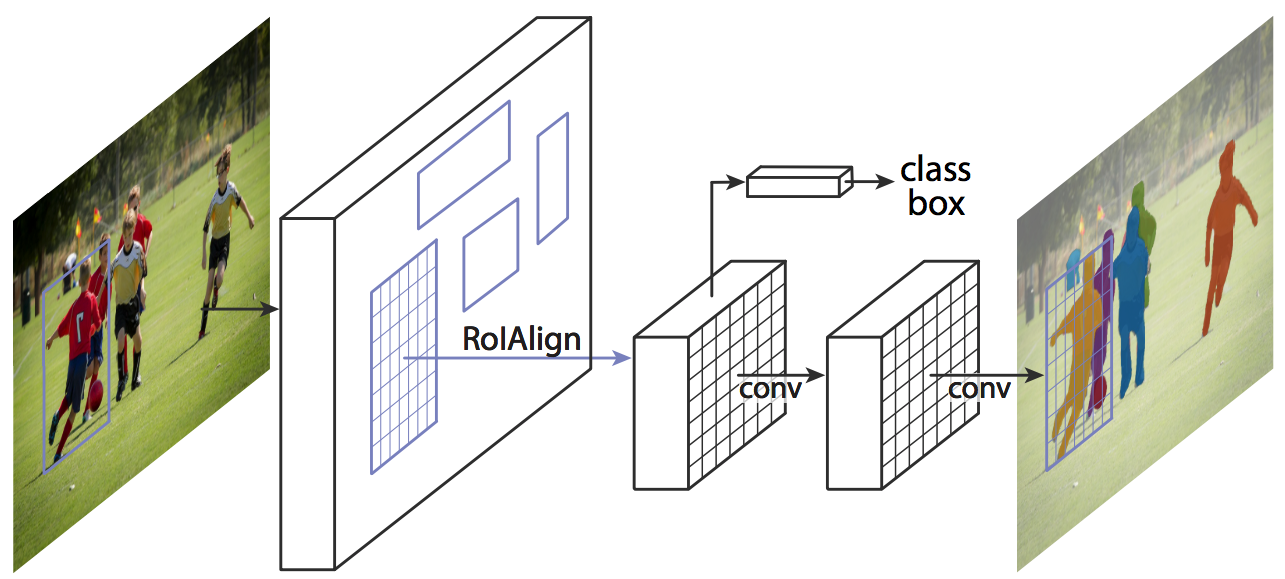
3. Mask Branch
addition of a parallel mask branch for predicting segmentation masks on each RoI → parallel predictions of (1) object mask, (2) class labels, and (3) bounding box offset
- mask branch: a small FCN (Fully Convolutional Network) applied to each RoI that predicts a segmentation mask in pixel-to-pixel manner (extracts much finer spatial layout of an object)
- predict a binary mask for each class independently, relying on the network’s RoI classification branch to predict the category (in previous works, classification depended on mask predictions)
- decoupling mask and class prediction was essential for performance in instance segmentation (shown through experiments)
- only adds a small computational overhead → enables a fast system and rapid experimentation
Modules
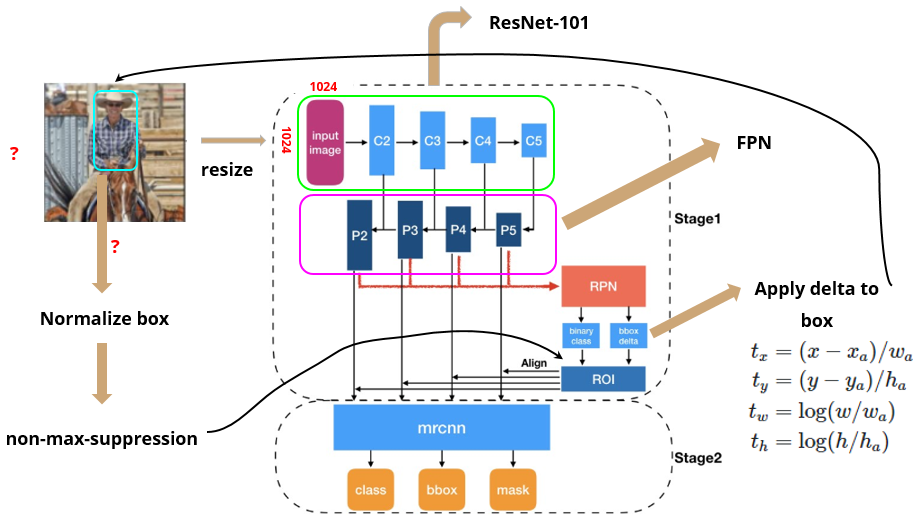
Recall: overall structure of Mask R-CNN
Backbone (part 1): ResNet-101
- CNN to extract features from raw images at low levels to higher levels
- input: 1024 x 1024 x 3 (RGB) image (resized)
- output: 32 x 32 x 2048 feature map → “C5” (1024/2/2/2/2/2 = 32)
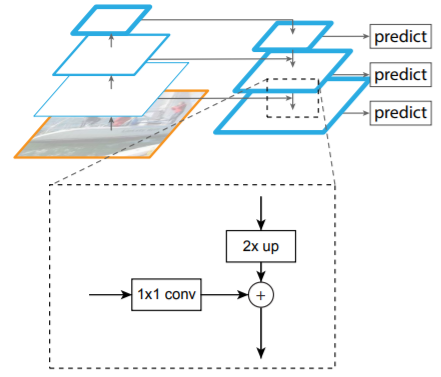
Backbone (part 2): Feature Pyramid Network
- bottom-up pathway → backbone (ResNet-101)
- top-bottom pathway: generate feature pyramid map with similar size to bottom-up pathway
- lateral connections: 1x1 conv to match #of filters in top-bottom pathway
[prev top-bottom layer]x2 + [curr bottom-up layer]*(1x1 conv) = [next top-bottom layer]
→ maintains strong semantic features at various resolution scales
- apply 3x3 conv before handing over to RPN (b/c feature data may be slightly jumbled up due to upsampling and addition)
Region Proposal Network
- lightweight NN that scans feature map in a sliding-window fashion → finds areas that contain objects
- input: feature maps from FPN
- RPN scans over feature map → anchors with 5 scales and 3 aspect ratios in total
- scans many anchors pretty fast (ex. 10ms in Faster R-CNN, a bit longer for Mask R-CNN due to larger image size and more anchors)
- sliding window handled by conv nature of RPN → scans all regions in parallel
- scan over feature map → reuse extracted features efficiently (by avoiding duplicate calculations)
- output:
- anchor class: foreground (FG, positive anchor) or background (BG) → FG class implies there is likely an object in that box
- bounding box refinement: refines box for positive anchor with delta (% change in x, y, w, h)
→ pick top anchors likely to contain objects and refine their location and size → if several anchors overlap: use NMS (Non-Max Suppression) to get final proposals (RoI)
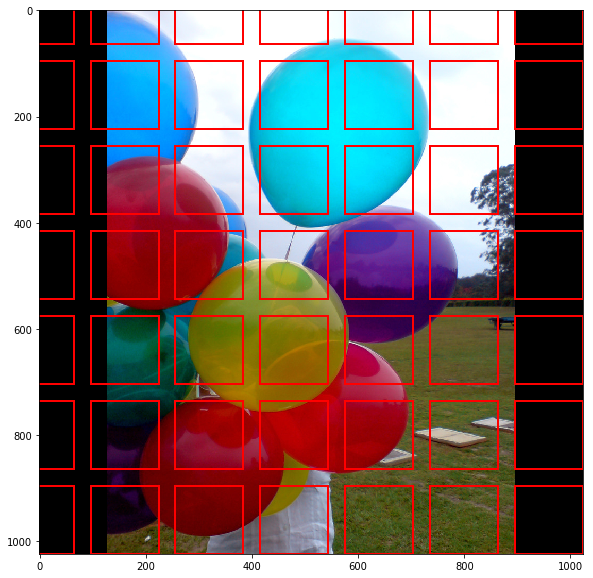
RPN is trained separately and does not share features with Mask R-CNN (but have same backbone → shareable)- RoIAlign to output
- needed because we need to hand over fixed input size to classifiers in the next step
- currently, RoI boxes have different sizes due to BB refinement
- use RoIAlign to crop part of feature map and resize it to a fixed size output (final proposals = RoI)
RoI Classifier & Bounding Box Regressor
- input: fixed size RoIs proposed by RPN
- output:
- Class of object in RoI: can classify regions to specific classes (ex. person, horse, chair, …) or background (→ RoI is discarded in this case)
- Bounding Box refinement: similar to RPN but further refines location and size of BB
Segmentation Masks
- conv network with input as positive regions selected by RoI Classifier
- (parallel just to bounding-box regression, and not classification…?.?) → parallel in backpropagation step! (get classification result in forward step and use that class for mask segmentation in parallel to the other branches in backprop step)

example of input image

example of segmentation masks for example input
- output: 28 x 28 low-res pixel masks for input → soft masks (represented with float numbers: hold more details than binary masks)
- in training: GT masks scaled down to 28 x 28 to compute the loss
- in inference: scale up predicted mask to the size of RoI BB → get final masks, one per object
- scale up using skimage.transform.resize() to perform bilinear interpolation
- do we have the GT mask? (binary) in COCO dataset?
- is GT mask scaled down in training by bilinear interpolation?
- is bilinear interpolation an invertible map?
# code to scale up predicted mask (28 x 28) into size of original input image def **unmold_mask**(mask, bbox, image_shape): """Converts a mask generated by the neural network to a format similar to its original shape. mask: [height, width] of type float. A small, typically 28x28 mask. bbox: [y1, x1, y2, x2]. The box to fit the mask in. Returns a binary mask with the same size as the original image. """ threshold = 0.5 y1, x1, y2, x2 = bbox mask = **resize**(mask, (y2 - y1, x2 - x1)) mask = np.where(mask >= threshold, 1, 0).astype(np.bool) # Put the mask in the right location. full_mask = np.zeros(image_shape[:2], dtype=np.bool) full_mask[y1:y2, x1:x2] = mask return full_mask def **resize**(image, output_shape, order=1, mode='constant', cval=0, clip=True, preserve_range=False, anti_aliasing=False, anti_aliasing_sigma=None): """A wrapper for Scikit-Image resize(). Scikit-Image generates warnings on every call to resize() if it doesn't receive the right parameters. The right parameters depend on the version of skimage. This solves the problem by using different parameters per version. And it provides a central place to control resizing defaults. """ if LooseVersion(skimage.__version__) >= LooseVersion("0.14"): # New in 0.14: anti_aliasing. Default it to False for backward # compatibility with skimage 0.13. return skimage.transform.resize( image, output_shape, order=order, mode=mode, cval=cval, clip=clip, preserve_range=preserve_range, anti_aliasing=anti_aliasing, anti_aliasing_sigma=anti_aliasing_sigma) else: return skimage.transform.resize( image, output_shape, order=order, mode=mode, cval=cval, clip=clip, preserve_range=preserve_range) - skimage.transform.resize(input image, output_shape): resizes image to match a certain output_shape size by interpolation to up-size or down-size N-dimensional images
- order of spline interpolation: 1 (bilinear)
- mode: constant, cval = 0 → points outside the boundaries of the input are filled with a constant value 0
- clip = True → clip output to range of values of the input image (0~255?)
- preserve_range = False → input image is converted
-
implementation code
def resize(image, output_shape, order=None, mode='reflect', cval=0, clip=True, preserve_range=False, anti_aliasing=None, anti_aliasing_sigma=None): """Resize image to match a certain size. Performs interpolation to up-size or down-size N-dimensional images. Note that anti-aliasing should be enabled when down-sizing images to avoid aliasing artifacts. For down-sampling with an integer factor also see `skimage.transform.downscale_local_mean`. Parameters ---------- image : ndarray Input image. output_shape : tuple or ndarray Size of the generated output image `(rows, cols[, ...][, dim])`. If `dim` is not provided, the number of channels is preserved. In case the number of input channels does not equal the number of output channels a n-dimensional interpolation is applied. Returns ------- resized : ndarray Resized version of the input. Other parameters ---------------- order : int, optional The order of the spline interpolation, default is 0 if image.dtype is bool and 1 otherwise. The order has to be in the range 0-5. See `skimage.transform.warp` for detail. mode : {'constant', 'edge', 'symmetric', 'reflect', 'wrap'}, optional Points outside the boundaries of the input are filled according to the given mode. Modes match the behaviour of `numpy.pad`. cval : float, optional Used in conjunction with mode 'constant', the value outside the image boundaries. clip : bool, optional Whether to clip the output to the range of values of the input image. This is enabled by default, since higher order interpolation may produce values outside the given input range. preserve_range : bool, optional Whether to keep the original range of values. Otherwise, the input image is converted according to the conventions of `img_as_float`. Also see https://scikit-image.org/docs/dev/user_guide/data_types.html anti_aliasing : bool, optional Whether to apply a Gaussian filter to smooth the image prior to down-scaling. It is crucial to filter when down-sampling the image to avoid aliasing artifacts. If input image data type is bool, no anti-aliasing is applied. anti_aliasing_sigma : {float, tuple of floats}, optional Standard deviation for Gaussian filtering to avoid aliasing artifacts. By default, this value is chosen as (s - 1) / 2 where s is the down-scaling factor, where s > 1. For the up-size case, s < 1, no anti-aliasing is performed prior to rescaling. Notes ----- Modes 'reflect' and 'symmetric' are similar, but differ in whether the edge pixels are duplicated during the reflection. As an example, if an array has values [0, 1, 2] and was padded to the right by four values using symmetric, the result would be [0, 1, 2, 2, 1, 0, 0], while for reflect it would be [0, 1, 2, 1, 0, 1, 2]. Examples -------- >>> from skimage import data >>> from skimage.transform import resize >>> image = data.camera() >>> resize(image, (100, 100)).shape (100, 100) """ image, output_shape = _preprocess_resize_output_shape(image, output_shape) input_shape = image.shape if image.dtype == np.float16: image = image.astype(np.float32) if anti_aliasing is None: anti_aliasing = not image.dtype == bool if image.dtype == bool and anti_aliasing: warn("Input image dtype is bool. Gaussian convolution is not defined " "with bool data type. Please set anti_aliasing to False or " "explicitely cast input image to another data type. Starting " "from version 0.19 a ValueError will be raised instead of this " "warning.", FutureWarning, stacklevel=2) factors = (np.asarray(input_shape, dtype=float) / # array([28,28,3]) np.asarray(output_shape, dtype=float)) # array([1024,1024,3]) # get array([0.02734375, 0.02734375, 1. ]) # Translate modes used by np.pad to those used by scipy.ndimage ndi_mode = _to_ndimage_mode(mode) # get 'constant' if anti_aliasing: # skip this stage for us, because we have anti_aliasing=None if anti_aliasing_sigma is None: anti_aliasing_sigma = np.maximum(0, (factors - 1) / 2) else: anti_aliasing_sigma = \ np.atleast_1d(anti_aliasing_sigma) * np.ones_like(factors) if np.any(anti_aliasing_sigma < 0): raise ValueError("Anti-aliasing standard deviation must be " "greater than or equal to zero") elif np.any((anti_aliasing_sigma > 0) & (factors <= 1)): warn("Anti-aliasing standard deviation greater than zero but " "not down-sampling along all axes") image = ndi.gaussian_filter(image, anti_aliasing_sigma, cval=cval, mode=ndi_mode) if NumpyVersion(scipy.__version__) >= '1.6.0': # The grid_mode kwarg was introduced in SciPy 1.6.0 order = _validate_interpolation_order(image.dtype, order) # get 1 b/c image.dtype is not binary zoom_factors = [1 / f for f in factors] # get [36.57142857142857, 36.57142857142857, 1.0] if order > 0: image = convert_to_float(image, preserve_range) # convert to float in [0,1] out = ndi.zoom(image, zoom_factors, order=order, mode=ndi_mode, cval=cval, grid_mode=True) # image size increased by 36.75배 # area beyond edges, filled with 0 (cval) # zoom: _clip_warp_output(image, out, order, mode, cval, clip) # clip output image to range of values of input image [0,1] # output: # TODO: Remove the fallback code below once SciPy >= 1.6.0 is required. # 2-dimensional interpolation # (like bilinear interpolation that we need!) elif len(output_shape) == 2 or (len(output_shape) == 3 and output_shape[2] == input_shape[2]): rows = output_shape[0] cols = output_shape[1] input_rows = input_shape[0] input_cols = input_shape[1] if rows == 1 and cols == 1: tform = AffineTransform(translation=(input_cols / 2.0 - 0.5, input_rows / 2.0 - 0.5)) else: # 3 control points necessary to estimate exact AffineTransform src_corners = np.array([[1, 1], [1, rows], [cols, rows]]) - 1 dst_corners = np.zeros(src_corners.shape, dtype=np.double) # take into account that 0th pixel is at position (0.5, 0.5) dst_corners[:, 0] = factors[1] * (src_corners[:, 0] + 0.5) - 0.5 dst_corners[:, 1] = factors[0] * (src_corners[:, 1] + 0.5) - 0.5 tform = AffineTransform() tform.estimate(src_corners, dst_corners) # Make sure the transform is exactly metric, to ensure fast warping. tform.params[2] = (0, 0, 1) tform.params[0, 1] = 0 tform.params[1, 0] = 0 out = warp(image, tform, output_shape=output_shape, order=order, mode=mode, cval=cval, clip=clip, preserve_range=preserve_range) else: # n-dimensional interpolation order = _validate_interpolation_order(image.dtype, order) coord_arrays = [factors[i] * (np.arange(d) + 0.5) - 0.5 for i, d in enumerate(output_shape)] coord_map = np.array(np.meshgrid(*coord_arrays, sparse=False, indexing='ij')) image = convert_to_float(image, preserve_range) out = ndi.map_coordinates(image, coord_map, order=order, mode=ndi_mode, cval=cval) _clip_warp_output(image, out, order, mode, cval, clip) return out-
ndimage.zoom
def zoom(input, zoom, output=None, order=3, mode='constant', cval=0.0, prefilter=True, *, grid_mode=False): # input = 28x28 mask, zoom = zoom_factors = [36.57142857142857, 36.57142857142857, 1.0] """ Zoom an array. The array is zoomed using spline interpolation of the requested order. Parameters ---------- %(input)s zoom : float or sequence The zoom factor along the axes. If a float, `zoom` is the same for each axis. If a sequence, `zoom` should contain one value for each axis. %(output)s order : int, optional The order of the spline interpolation, default is 3. The order has to be in the range 0-5. %(mode_interp_constant)s %(cval)s %(prefilter)s grid_mode : bool, optional If False, the distance from the pixel centers is zoomed. Otherwise, the distance including the full pixel extent is used. For example, a 1d signal of length 5 is considered to have length 4 when `grid_mode` is False, but length 5 when `grid_mode` is True. See the following visual illustration: .. code-block:: text | pixel 1 | pixel 2 | pixel 3 | pixel 4 | pixel 5 | |<-------------------------------------->| vs. |<----------------------------------------------->| The starting point of the arrow in the diagram above corresponds to coordinate location 0 in each mode. Returns ------- zoom : ndarray The zoomed input. Notes ----- For complex-valued `input`, this function zooms the real and imaginary components independently. .. versionadded:: 1.6.0 Complex-valued support added. Examples -------- >>> from scipy import ndimage, misc >>> import matplotlib.pyplot as plt >>> fig = plt.figure() >>> ax1 = fig.add_subplot(121) # left side >>> ax2 = fig.add_subplot(122) # right side >>> ascent = misc.ascent() >>> result = ndimage.zoom(ascent, 3.0) >>> ax1.imshow(ascent, vmin=0, vmax=255) >>> ax2.imshow(result, vmin=0, vmax=255) >>> plt.show() >>> print(ascent.shape) (512, 512) >>> print(result.shape) (1536, 1536) """ if order < 0 or order > 5: raise RuntimeError('spline order not supported') input = numpy.asarray(input) if input.ndim < 1: raise RuntimeError('input and output rank must be > 0') zoom = _ni_support._normalize_sequence(zoom, input.ndim) output_shape = tuple( [int(round(ii * jj)) for ii, jj in zip(input.shape, zoom)]) complex_output = numpy.iscomplexobj(input) output = _ni_support._get_output(output, input, shape=output_shape, complex_output=complex_output) if complex_output: # import under different name to avoid confusion with zoom parameter from scipy.ndimage.interpolation import zoom as _zoom kwargs = dict(order=order, mode=mode, prefilter=prefilter) _zoom(input.real, zoom, output=output.real, cval=numpy.real(cval), **kwargs) _zoom(input.imag, zoom, output=output.imag, cval=numpy.imag(cval), **kwargs) return output if prefilter and order > 1: padded, npad = _prepad_for_spline_filter(input, mode, cval) filtered = spline_filter(padded, order, output=numpy.float64, mode=mode) else: npad = 0 filtered = input if grid_mode: # warn about modes that may have surprising behavior suggest_mode = None if mode == 'constant': suggest_mode = 'grid-constant' elif mode == 'wrap': suggest_mode = 'grid-wrap' if suggest_mode is not None: warnings.warn( ("It is recommended to use mode = {} instead of {} when " "grid_mode is True." ).format(suggest_mode, mode) ) mode = _ni_support._extend_mode_to_code(mode) zoom_div = numpy.array(output_shape) zoom_nominator = numpy.array(input.shape) if not grid_mode: zoom_div -= 1 zoom_nominator -= 1 # Zooming to infinite values is unpredictable, so just choose # zoom factor 1 instead zoom = numpy.divide(zoom_nominator, zoom_div, out=numpy.ones_like(input.shape, dtype=numpy.float64), where=zoom_div != 0) zoom = numpy.ascontiguousarray(zoom) _nd_image.zoom_shift(filtered, zoom, None, output, order, mode, cval, npad, grid_mode) return output
-
Loss
Multi-task loss on each sampled RoI: $L =$ $L_{cls} + L_{box}$ $+ L_{mask}$
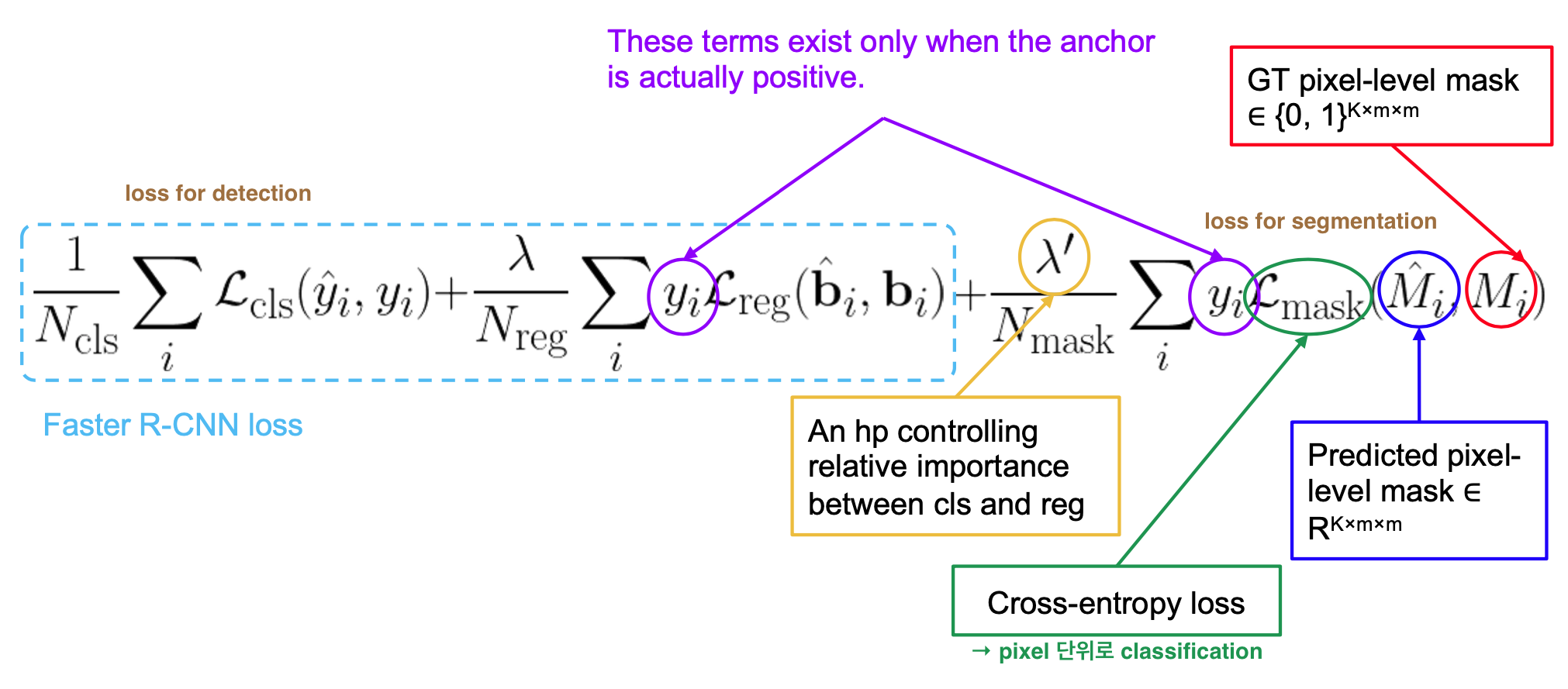
ref) 2021-1 MLVU Lecture 10, slide 23
- $L_{cls}$ and $L_{box}$: identical to Fast R-CNN
- $L_{mask}$ = average binary cross-entropy loss [ per-pixel sigmoid(output) ] ← for positive regions selected by RoI Classifier
- output: $Km^2$ -dimensional output for each RoI ← encodes K binary masks of resolution m x m (one for each of K classes)
- RoI associated with GT class k → $L_{mask}$ only defined on k-th mask (other mask outputs do not contribute to the loss)
- allows network to generate masks for every class without competition among classes, but rely on the dedicated classification branch to predict the class label used to select the output mask → decouples mask and class prediction (?) → (whereas FCNs usually perform per-pixel multi-class categorization → couples segmentation & classification)
Results
- data: COCO
- train with 80k train images and 35k subset of validation images (5k val for ablation studies)
- evaluation metric: AP(Average Precision, averaged over IoU thresholds) using mask IoU → use recall values as thresholds
- tradeoff between recall and precision as threshold value changes ($\approx$ AUC)
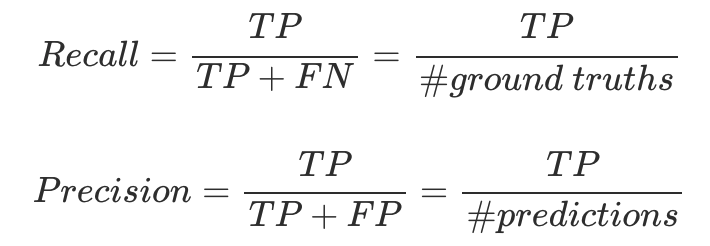
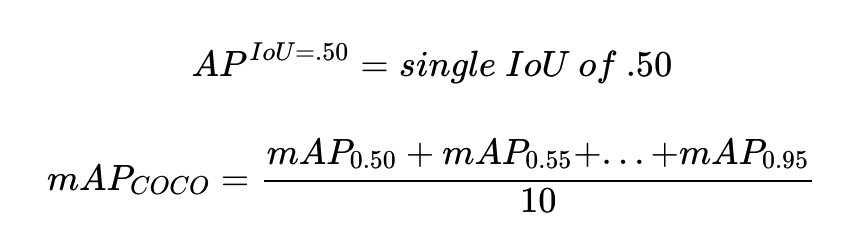
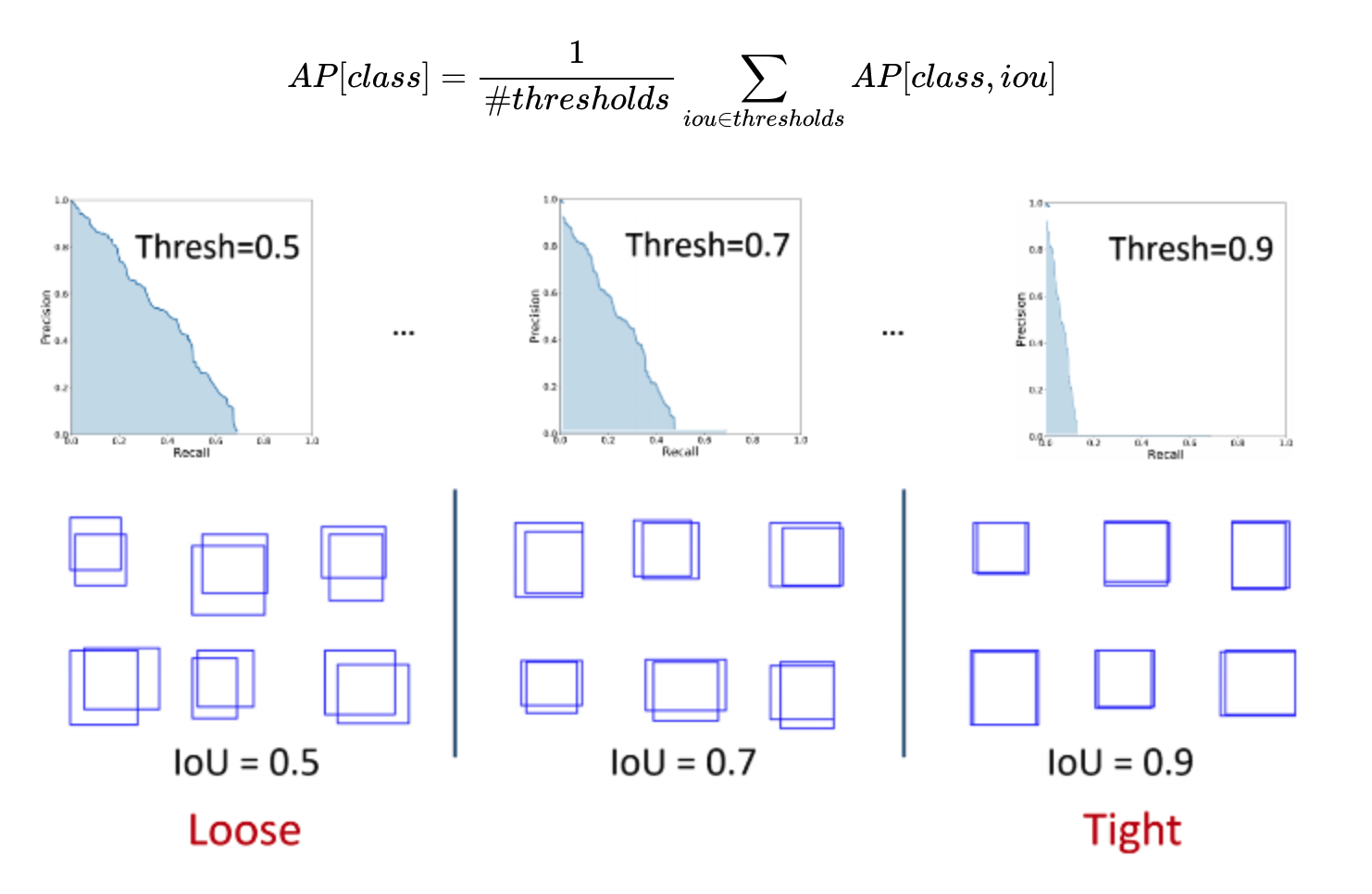 → and then average APs of all classes
→ and then average APs of all classes
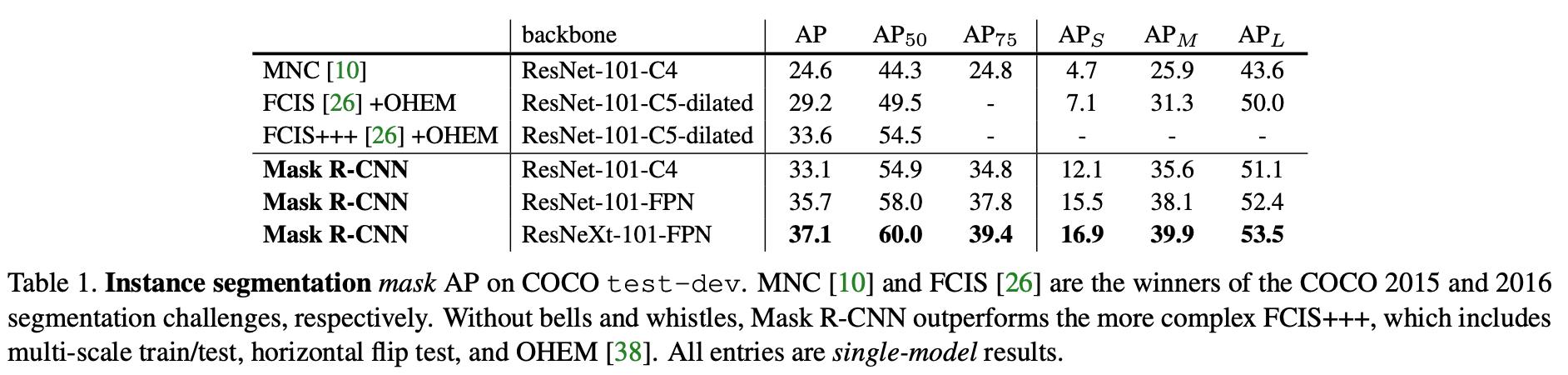
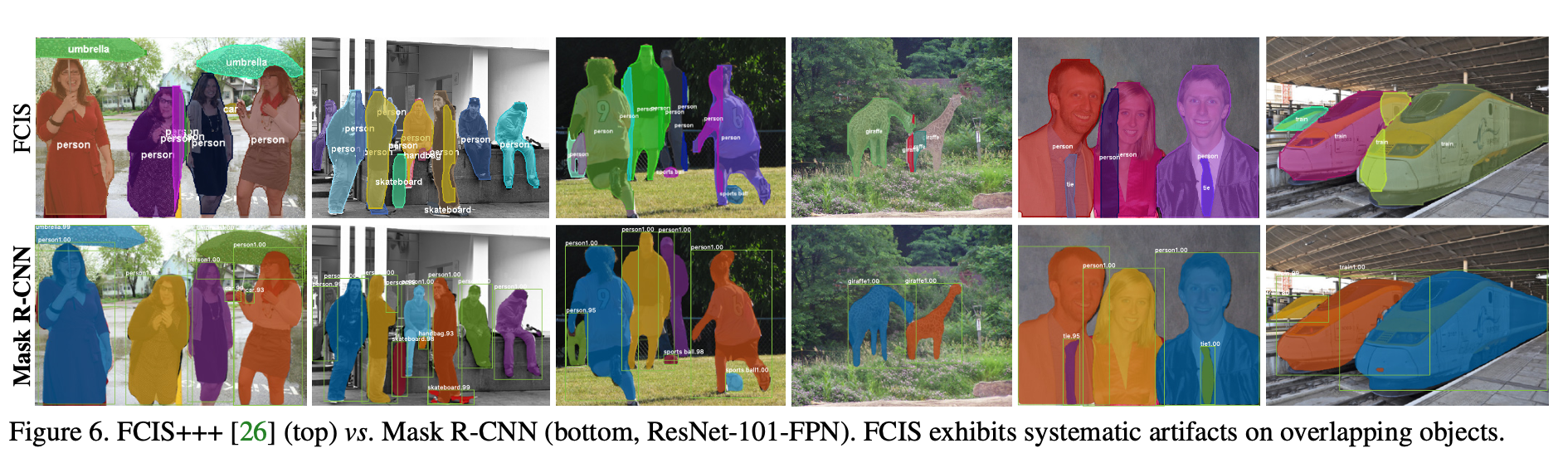
- outperforms previous SOTA models in terms of AP, and in challenging conditions (ex. overlapping instances)
- Ablation (reasoning for structure of Mask R-CNN model)→ an explanation for why each individual idea is useful
- Multinomial vs. independent masks using per-pixel softmax and multinomial loss (commonly used in FCN): independent masks win → decoupling of mask and class prediction tasks
- Class-specific vs. class-agnostic masks: class-specific masks win (one m x m mask per class)
- RoIAlign vs. RoIPool vs. RoIWarp: RoIAlign wins (AP ↑ by 3pts than RoIPool $\approx$ RoIWarp) → RoIAlign is insensitive to max/average pool → RoIPool, RoIWarp: both quantizes RoI, losing alignment with the input → proper alignment is key!
- Timing
ResNet-101-C4 variant takes ∼400ms as it has a heavier box head (Figure 4), so we do not recommend using the C4 variant in practice.- fast, but can be optimized for speed, and better speed/accuracy tradeoffs (e.g. by varying image sizes and proposal numbers)
- fast training: ResNet-101-FPN on COCO trainval35k takes 44 hrs in synchronized 8-GPU implementation
- Generality & Flexibility of Mask R-CNN
- human pose estimation: model a keypoint’s location as a one-hot mask → adopt Mask R-CNN to predict K masks, one for each K keypoint types (ex. left shoulder, right elbow)
References
https://firiuza.medium.com/roi-pooling-vs-roi-align-65293ab741db
https://erdem.pl/2020/02/understanding-region-of-interest-part-2-ro-i-align
https://jonathan-hui.medium.com/map-mean-average-precision-for-object-detection-45c121a31173
https://www.sciencedirect.com/topics/engineering/bilinear-interpolation
https://ganghee-lee.tistory.com/39 (GAP vs. FCN)
https://ganghee-lee.tistory.com/41 (ResNet)
https://ganghee-lee.tistory.com/37 (Faster R-CNN)
https://github.com/Kanghee-Lee/Faster-RCNN_TF-RPN-/blob/master/RPN.ipynb (Faster R-CNN)
https://github.com/matterport/Mask_RCNN/blob/master/mrcnn/model.py
https://ganghee-lee.tistory.com/40 (Mask R-CNN)
https://scikit-image.org/docs/dev/api/skimage.transform.html#skimage.transform.resize
